Partial dates are very common in clinical trials, such as AE that allow some parts of the date or time to be missing. However, when you create the ADaM dataset for AE, some variables like ASTDT (Analysis Start Date) or AENDT (Analysis End Date) are numeric, so they can be derived only when the date is complete and then you can calculate the durations.
In the trials, we would actually draw up a plan to define the rules for how to impute the partial date. But here, I simplify the imputation rule as shown below to illustrate its implementation in R and SAS:
- If the day of analysis start date is missing then impute the first day of the month. If both the day and month are missing then impute to 01-Jan.
- If the day of analysis end date is missing then impute the last day of the month. If both the day and month are missing then impute to 31-Dec.
- If the imputed analysis end date is after the last alive date then set it to the last alive date.
Manipulating in SAS
Firstly, let’s create dummy data in SAS that includes four variables.
data dummy;
length USUBJID $20. LSTALVDT $20. AESTDTC $20. AEENDTC $20.;
input USUBJID $ LSTALVDT $ AESTDTC $ AEENDTC $;
datalines;
SITE01-001 2023-01-10 2019-06-18 2019-06-29
SITE01-001 2023-01-10 2020-01-02 2020-02
SITE01-001 2023-01-10 2022-03 2022-03
SITE01-001 2023-01-10 2022-06 2022-06
SITE01-001 2023-01-10 2023 2023
;
run;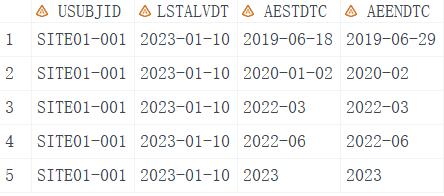
USUBJID, unique subject identifier.LSTALVDT, last known alive date.AESTDTC, start date of adverse event.AEENDTC, end date of adverse event.
And we can see from the above rules that concatenating "01" with the date that misses the day is very easy. However if we want to calculate the AENDT, we need to consider which day is matched with each month, for example, the 28th or 29th, 30th or 31th. So we need to apply the intnx function to get the last day correctly.
data dummy_2;
set dummy;
if length(AESTDTC)=7 then do;
ASTDTF="D";
ASTDT=catx('-', AESTDTC, "01");
end;
else if length(AESTDTC)=4 then do;
ASTDTF="M";
ASTDT=catx('-', AESTDTC, "01-01");
end;
else if length(AESTDTC)=10 then ASTDT=AESTDTC;
if length(AEENDTC)=7 then do;
AENDTF="D";
AEENDTC_=catx('-', AEENDTC, "01");
AENDT=put(intnx('month', input(AEENDTC_,yymmdd10.), 0, 'E'), yymmdd10.);
end;
else if length(AEENDTC)=4 then do;
AENDTF="M";
AENDT=catx('-', AEENDTC, "12-31");
end;
else if length(AEENDTC)=10 then AENDT=AEENDTC;
if input(AENDT,yymmdd10.)>input(LSTALVDT,yymmdd10.) then AENDT=LSTALVDT;
drop AEENDTC_;
run;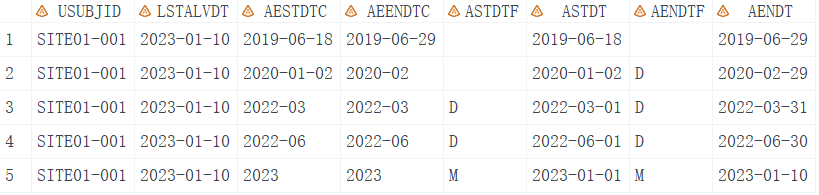
From the output we can see that when the day of date is missing, we set the imputation flag to 'D' as the flag variable, like ASTDTF. If the month of the date is missing, set it to "M". It also considers leap years and sets the date to the last alive date if the imputed date is later than the last alive date. So I suppose all the dates have been imputed correctly.
Manipulating in R
Then let’s create the same dummy to see how to conduct the rules in R.
library(tidyverse)
library(lubridate)
dummy <- tibble(
USUBJID = "SITE01-001",
LSTALVDT = "2023-01-10",
AESTDTC = c("2019-06-18", "2020-01-02", "2022-03", "2022-06", "2023"),
AEENDTC = c("2019-06-29", "2020-02", "2022-03", "2022-06", "2023")
)The dummy data can be shown below.
# A tibble: 5 × 4
USUBJID LSTALVDT AESTDTC AEENDTC
<chr> <chr> <chr> <chr>
1 SITE01-001 2023-01-10 2019-06-18 2019-06-29
2 SITE01-001 2023-01-10 2020-01-02 2020-02
3 SITE01-001 2023-01-10 2022-03 2022-03
4 SITE01-001 2023-01-10 2022-06 2022-06
5 SITE01-001 2023-01-10 2023 2023 And then we follow the rules as the SAS used to impute the partial date in R. To get the last day of each month's imputation, we'd better use the rollback() and ceiling_date() functions in the lubridate package to get the correct day considering the leap years. In addition, others are the common functions in the tidyverse package to manipulate the data, like case_when() and select().
dummy_2 <- dummy %>%
mutate(
ASTDTF = case_when(
str_length(AESTDTC) == 4 ~ "M",
str_length(AESTDTC) == 7 ~ "D"
),
ASTDT_ = case_when(
str_length(AESTDTC) == 4 ~ str_c(AESTDTC, "01-01", sep = "-"),
str_length(AESTDTC) == 7 ~ str_c(AESTDTC, "01", sep = "-"),
is.na(ASTDTF) ~ AESTDTC
),
ASTDT = ymd(ASTDT_),
AENDTF = case_when(
str_length(AEENDTC) == 4 ~ "M",
str_length(AEENDTC) == 7 ~ "D"
),
AENDT_ = case_when(
str_length(AEENDTC) == 4 ~ str_c(AEENDTC, "12-31", sep = "-"),
str_length(AEENDTC) == 7 ~ str_c(AEENDTC, "-15"),
is.na(AENDTF) ~ AEENDTC
),
AENDT = case_when(
str_length(AEENDTC) == 7 ~ rollback(ceiling_date(ymd(AENDT_), "month")),
TRUE ~ ymd(AENDT_)
),
AENDT = if_else(AENDT > ymd(LSTALVDT), ymd(LSTALVDT), AENDT)
) %>%
select(-ASTDT_, -AENDT_)Here we can see that the output is consistent with the SAS. It's very easy in R, right? You can also use many useful functions to transfer the different date types, for example from date9. to yymmdd10. like dmy("01Jan2023"). Honestly the lubridate package can provide a series of functions to deal with date manipulation, such as using interval() to calculate the duration of AEs.
# A tibble: 5 × 8
USUBJID LSTALVDT AESTDTC AEENDTC ASTDTF ASTDT AENDTF AENDT
<chr> <chr> <chr> <chr> <chr> <date> <chr> <date>
1 SITE01-001 2023-01-10 2019-06-18 2019-06-29 NA 2019-06-18 NA 2019-06-29
2 SITE01-001 2023-01-10 2020-01-02 2020-02 NA 2020-01-02 D 2020-02-29
3 SITE01-001 2023-01-10 2022-03 2022-03 D 2022-03-01 D 2022-03-31
4 SITE01-001 2023-01-10 2022-06 2022-06 D 2022-06-01 D 2022-06-30
5 SITE01-001 2023-01-10 2023 2023 M 2023-01-01 M 2023-01-10Using admiral Package
Maybe you would say if there is a package that can deal with date imputation for ADaM. A manipulation structure that is wrapped in a series of functions to sort out the common imputation situations in ADaM. There's no doubt that you can believe the admiral package. Let me show some examples here to demonstrate how to use it for imputing partial dates.
library(admiral)
dummy %>%
derive_vars_dt(
dtc = AESTDTC,
new_vars_prefix = "AST",
highest_imputation = "M",
date_imputation = "first"
) %>%
mutate(LSTALVDT = ymd(LSTALVDT)) %>%
derive_vars_dt(
dtc = AEENDTC,
new_vars_prefix = "AEND",
highest_imputation = "M",
date_imputation = "last",
max_dates = vars(LSTALVDT)
)Isn't the code quite straightforward? If your date vector is date time (DTM), you can use derive_vars_dtm() instead.
# A tibble: 5 × 8
USUBJID LSTALVDT AESTDTC AEENDTC ASTDT ASTDTF AENDDT AENDDTF
<chr> <date> <chr> <chr> <date> <chr> <date> <chr>
1 SITE01-001 2023-01-10 2019-06-18 2019-06-29 2019-06-18 NA 2019-06-29 NA
2 SITE01-001 2023-01-10 2020-01-02 2020-02 2020-01-02 NA 2020-02-29 D
3 SITE01-001 2023-01-10 2022-03 2022-03 2022-03-01 D 2022-03-31 D
4 SITE01-001 2023-01-10 2022-06 2022-06 2022-06-01 D 2022-06-30 D
5 SITE01-001 2023-01-10 2023 2023 2023-01-01 M 2023-01-10 MI'm planning to learn how to use the admiral package, for example, by building ADaM ADRS. I suppose this package improves the ecology of R greatly in drug trials.
Reference
Common Dating in R: With an example of partial date imputation
Tips to Manipulate the Partial Dates
Date and Time Imputation